In a previous article I showed how to visualise the results of a classifier using ggplot2 in R. In the same article I mentioned that Alex, a colleague at Forward, had suggested looking further at R’s caret package that would produce more detailed statistics about the overall performance of the classifer and within individual classes.
Confusion Matrix
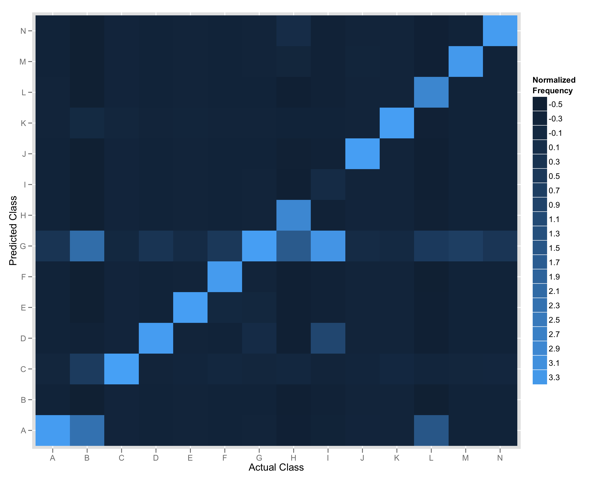
Using ggplot2 we can produce a plot like the one below: a visual representation of a confusion matrix. It gives us a nice overview but doesn’t reveal much about the specific performance characteristics of our classifier.
To produce our measures, we run our classifier across a set of test data and capture both the actual class and the predicted class. Our results are stored in a CSV file and will look a little like this:
actual, predicted A, B B, B, C, C B, A
Analysing with Caret
With our results data as above we can run the following to produce a confusion matrix with caret:
results.matrix now contains a confusionMatrix full of information. Let’s take a look at some of what it shows. The first table shows the contents of our matrix:
Reference Prediction A B C D A 211 3 1 0 B 9 26756 6 17 C 1 12 1166 1 D 0 18 3 1318
Each column holds the reference (or actual) data and within each row is the prediction. The diagonal represents instances where our observation correctly predicted the class of the item.
The next section contains summary statistics for the results:
Overall Statistics
Accuracy : 0.9107
95% CI : (0.9083, 0.9131)
No Information Rate : 0.5306
P-Value [Acc > NIR] : < 2.2e-16Overall accuracy is calculated at just over 90% with a p-value of 2 x 10^-16, or 0.00000000000000022. Our classifier seems to be doing a pretty reasonable job of classifying items.
Our classifier is being tested by putting items into 1 of 13 categories- caret also produces a final section of statistics for the performance of each class.
Class: A Class: B ... Class: J Sensitivity 0.761733 0.9478 0.456693 Specificity 0.998961 0.9748 0.999962 Pos Pred Value 0.793233 0.9770 0.966667 Neg Pred Value 0.998753 0.9429 0.998702 Prevalence 0.005206 0.5306 0.002387 Detection Rate 0.003966 0.5029 0.001090 Detection Prevalence 0.005000 0.5147 0.001128
The above shows some really interesting data.
Sensitivity and specificity respectively help us measure the performance of the classifier in correctly predicting the actual class of an item and not predicting the class of an item that is of a different class; it measures true positive and true negative performance.
From the above data we can see that our classifier correctly identified class B 94.78% of the time. That is, when we should have predicted class B we did. Further, when we shouldn’t have predicted class B we didn’t for 97.48% of examples. We can contrast this to class J: our specificity (true negative) is over 99% but our sensitivity (true positive) is around 45%; we do a poor job of positively identifying items of this class.
Caret has also calculated a prevalence measure- that is, of all observations, how many were of items that actually belonged to the specified class; it calculates the prevalence of a class within a population.
Using the previously defined sensitivity and specificity, and prevalance measures caret can calculate Positive predictive value and Negative predictive value. These are important as they reflect the probability that a true positive/true negative is correct given knowledge about the prevalence of classes within the population. Class J has a positive predictive value of over 96%: despite our classifier only being able to positively identify objects 45% of the time there’s a 96% chance that, when it does, such a classification is correct.
The caret documentation has some references to relevant papers discussing the measures it calculates.