Earlier in the year, myself and some colleagues started working on building better data processing tools for uSwitch.com. Part of the theory/reflection of this is captured in a presentation I was privileged to give at EuroClojure (titled Users as Data).
In the last few days, our data team (Thibaut, Paul and I) have been playing around with some of the data we collect and using it to build some classifiers. Precision and Recall provide quantitative measures but reading through Machine Learning for Hackers showed some nice ways to visualise results.
Binary Classifier
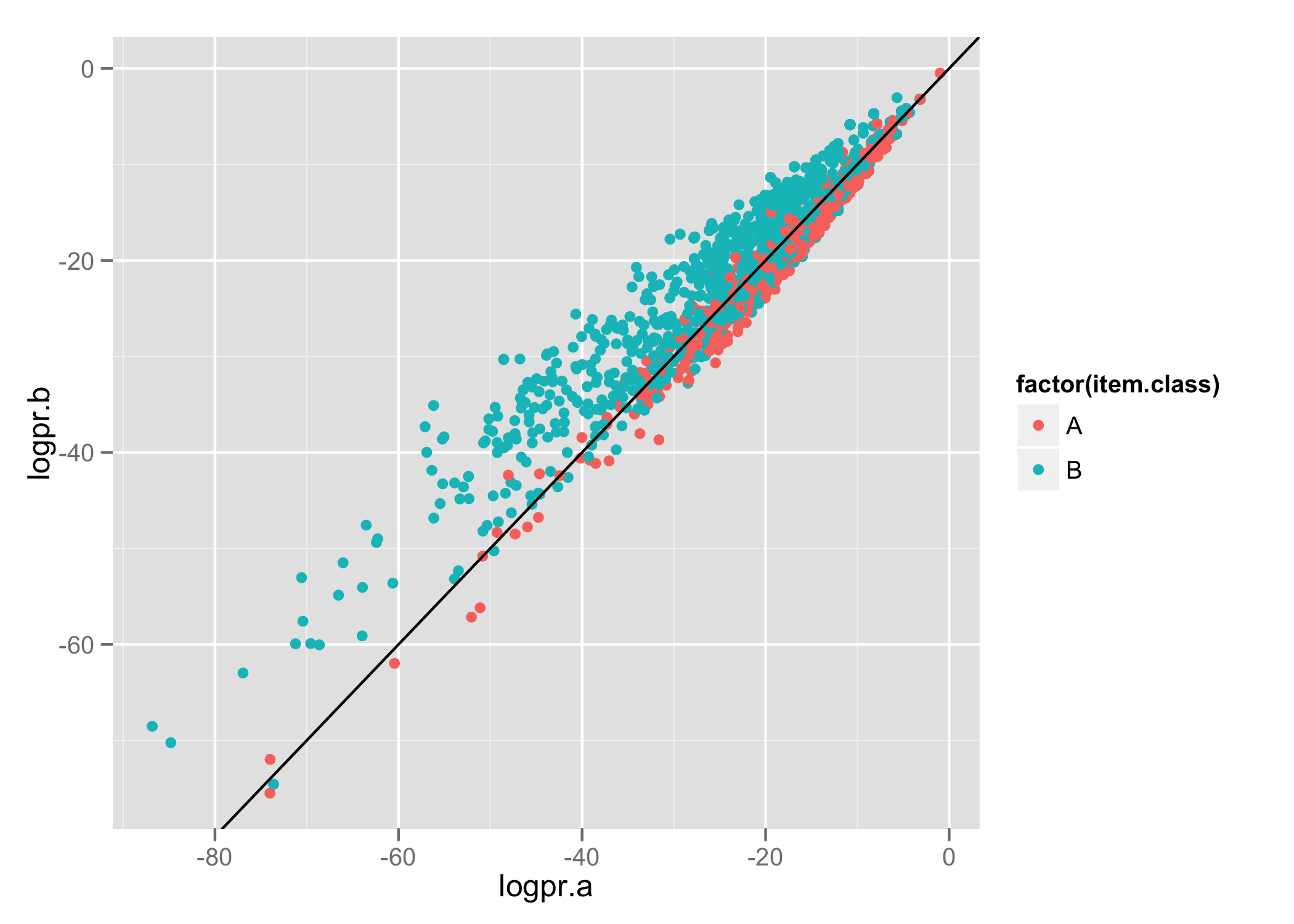
Our first classifier attempted to classify data into 2 groups. Using R and ggplot2 I produced a plot (similar to the one presented in the Machine Learning for Hackers book) to show the results of the classifier.
Our results were captured in a CSV file and looked a little like this:
A,0.25,0.15 A,0.2,0.19 B,0.19,0.25
Each line contains the item's actual class, the predicted probability for membership of class A, and the predicted probability for membership of class B. Using ggplot2 we produce the following:
Items have been classified into 2 groups- A and B. The axis show the log probability (we’re using Naive Bayes to classify items) that the item belongs to the specified class. We use colour to identify the actual class for items and draw a line to represent the decision boundary (i.e. which of the 2 classes did our model predict).
This lets us nicely see the relationship between predicted and actual classes.
We can see there’s a bit of an overlap down the decision boundary line and we’re able to do a better job for classifying items in category B than A.
The R code to produce the plot above is as follows. Note that because we had many millions of observations I randomly sampled to make it possible to compute on my laptop :)
More Classes!
But what if we want to see compare the results when we’re classifying items into more than 1 group?
After chatting to Alex Farquhar (another data guy at Forward) he suggested plotting a confusion matrix.
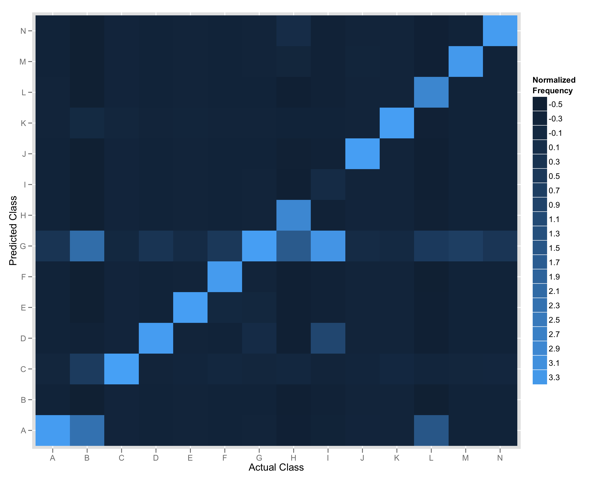
Below shows the plot we produced that compares the actual and predicted classes for 14 items.
The y-axis shows the predicted class for all items, and the x-axis shows the actual class. The tiles are coloured according to the frequency of the intersection of the two classes thus the diagonal represents where we predict the actual class. The colour represents the relative frequency of that observation in our data; given some classes occur more frequently we normalize the values before plotting.
Any row of tiles (save for the diagonal) represents instances where we falsely identified items as belonging to the specified class. In the rendered plot we can see that items in Class G were often identified for items belonging to all other classes.
Our input data looked a little like this:
1,0,0 0,3,0 1,0,2
It’s a direct encoding of our matrix- each column represents data for classes A to N, and each row represents data for classes A to N. The diagonal holds data for A,A, B,B, etc.
The R code to plot the confusion matrix is as follows:
Alex also suggested using the caret package which includes a function to build the confusion matrix from observations directly and also provides some useful summary statistics. I’m going to hack on our classifier’s Clojure code a little more and will be sure to post again with the findings!